Community Research and Assistance
RSD3 is available for requests from the research community for assistance in producing new specialized workflows and toolkits. The supported effort for this is limited, so we currently solicit short proposals for this type of work. More information will be forthcoming on the proposal process.
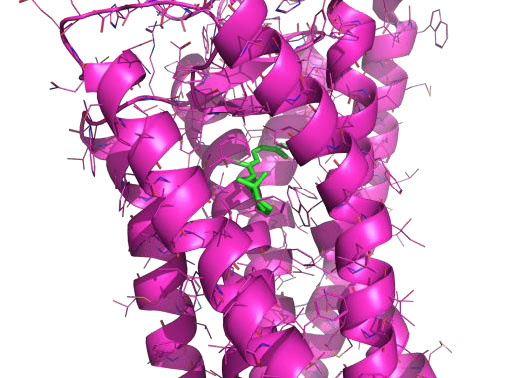
Natural products for the treatment of macular degeneration
Anne Hanneken, TSRI
Building on preliminary biochemical data connecting natural products with treatments for macular degeneration, this project characterized allosteric binding sites on human visual opsins, evaluated binding of existing compounds, and screened for new compounds, all with the long-term aim of stabilizing visual opsins/pigments for therapeutic benefit. A motivated graduate student in the laboratory used AutoSite and AutoDockVina to identify a new site on the surface of the receptor and dock several existing compounds that have shown biological activity. They are now interpreting these results to generate structural hypotheses for the interaction of the compounds with the retinal chromophore, and planning additional studies, including molecular dynamics simulations of the interaction and designing methods to improve binding and efficacy of the compounds.
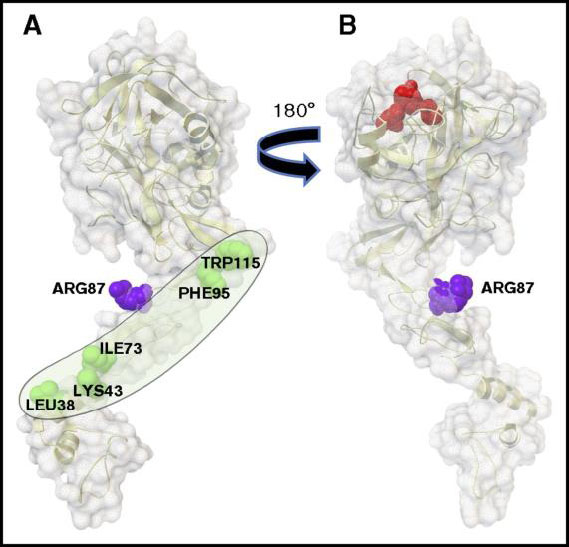
Modeling interactions of PAR1 with Activated Protein C (APC)
John Griffin, TSRI
The project built structural models of the N-terminus of Protease Activated Receptor 1 (PAR-1) interacting with Activated Protein C (APC) to identify mutations that will favor cleavage of PAR-1 at R46 over R41, as part of research related to blood clot prevention, neural protection in case of stroke, and sepsis protection. AutoDockFR and AutoDock CrankPep were used to dock peptides of various sizes (3 to 8 amino acids) around positions R41 and R46 of PAR-1. Subsequent homology modeling followed by molecular dynamics (MD) simulations were performed to model the interactions between the APC and PAR-1 segments. Based on these models, two mutations of APC were proposed, T254F and A195Q, that were subsequently tested. Additionally, MD simulations explored the structural fluctuation of the C-terminus of the APC light chain and its possible interactions with other segments. Despite these large fluctuations, the C-terminus of the APC light chain stays within the region interacting with Protein S. The conformation most often sampled during the MD simulation shows that K150 and K151 are in close contact with L73, which is also known to alter the binding with Protein S. On the other hand, R143, K146 and R147A are located above the K150 and K151 residues, i.e., further away from the membrane and closer to the protease domain of APC. This suggests that the interface between the APC light chain and Protein S is extended and has a boundary near the APC protease domain.
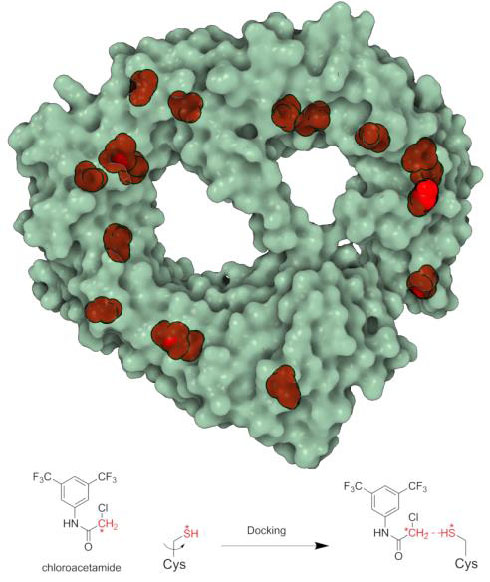
Proteome-wide covalent ligand discovery in native biological systems
Ben Cravatt, TSRI
This project provided structure-based insight to the results of a large proteomics screening. Fragment-based covalent ligand discovery was used to identify small molecule probes for proteins that have proven difficult to target using high throughput screening of complex compound libraries. His lab performed a quantitative analysis of cysteine-reactive small-molecule fragments screened against thousands of proteins in human proteomes and cells. Covalent ligands were identified for >700 cysteines found in both druggable proteins and proteins deficient in chemical probes, including transcription factors, adaptor/scaffolding proteins, and uncharacterized proteins. A new docking protocol (Reactive Docking) was developed specifically for this task. In silico fragment library containing all chloroacetamide and acrylamide fragments was prepared using the OpenBabel library with custom Python scripts. Three-dimensional coordinates were generated from SMILES strings, calculating their protonation state at pH 7.4, and then minimizing them using MMF94 s forcefield (50K iterations steepest descent; 90K conjugate gradient); for chiral molecules with undefined configuration, all enantiomers were generated, resulting in 53 total fragments. For each protein, the UniProtKB accession number was used to filter the PDB36. Structures determined by X-ray crystallography were selected, privileging higher sequence coverage and structure resolution. Fragments were modeled in their reactive form (that is, with explicit chloroacetamide and acrylamide warheads), and protein structures were prepared following the standard AutoDock protocol.
Design of binders of HIV-IN active site as HIV-1 drug candidates
Dmitry Lyumkis, Salk Institute
This project used thecryo-EM structure of HIV-1 intasome to drive the design of derivatives of known drugs to escape mutations leading to viral drug resistance. Given the high quality of the data sampled, his laboratory was able to resolve the position of a number of waters involved in the mediation of interactions between the ligands and the target protein. Structural data and docking experiments showed that these waters clearly play a role in modulating the ligand binding affinity. In collaboration with the Lyumkis lab, we set up a pipeline that combines docking protocols with subsequent molecular dynamics refinements. We ran MD computations to help rank the free energy of the docked compounds, which can now be used for compound acquisition and experimental testing.
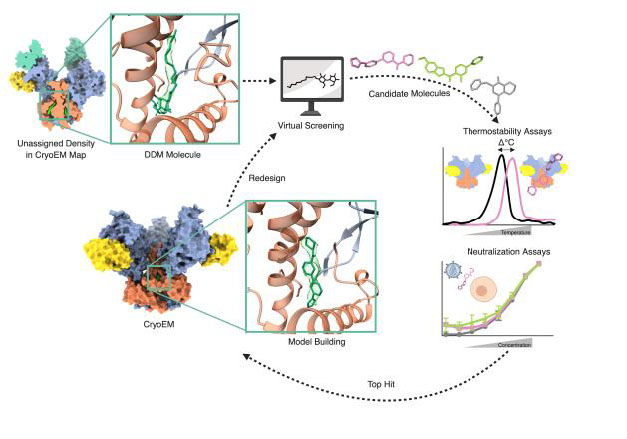
High-throughput virtual screening of potential binders of HIV-Env allosteric site
Andrew Ward, TSRI
This project used the cryo-EM structure of HIV-1 Env determined to screen for novel molecules capable of inhibiting the conformational change of Env upon binding to the host cell receptors. Using a preliminary structure that showed the presence of a co-solvent, a high throughput virtual screening was performed, screening a diversity set of the ChemBridge library (~70k molecules) using AutoDock Vina. The screening led to the identification of a series of hits sharing a common aromatic scaffold. One of these virtual hits was selected to be structurally resolved, showing that the docked coordinates matched very closely with the experimentally determined structure. The hits of this first iteration round were used to identify molecules with similar scaffolds, in order to find derivatives with higher affinity, screening ~130M molecules. The screening has been completed and the analysis on-going.